
Saccharomyces cerevisiae Protein-Protein Interaction Binding Sites Database
Introduction
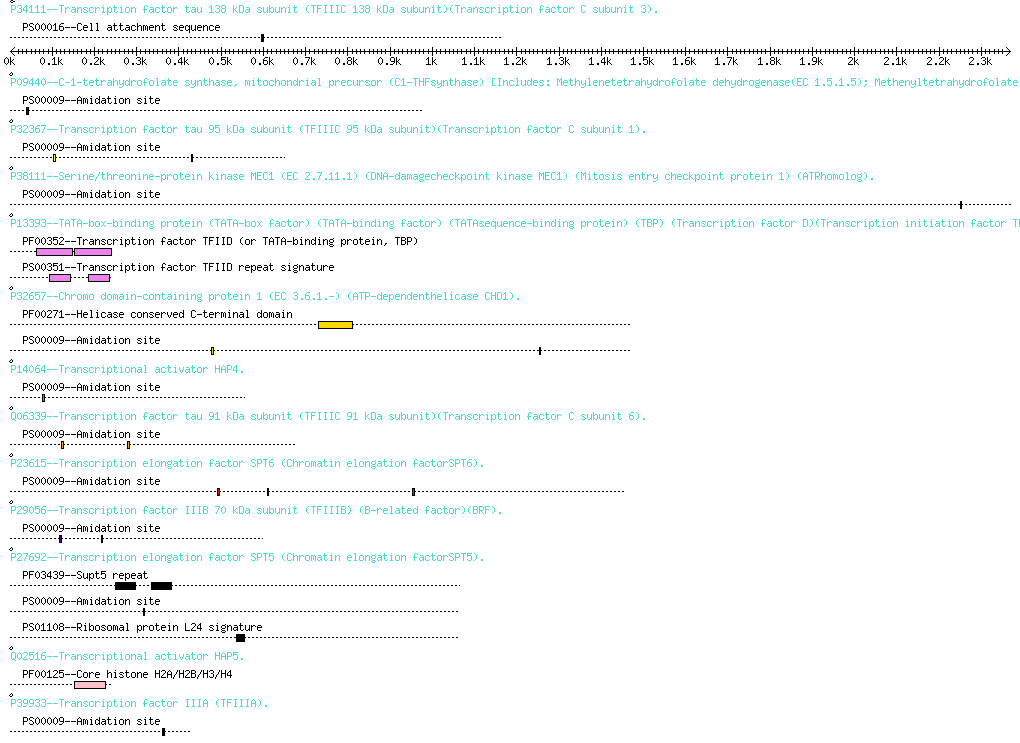
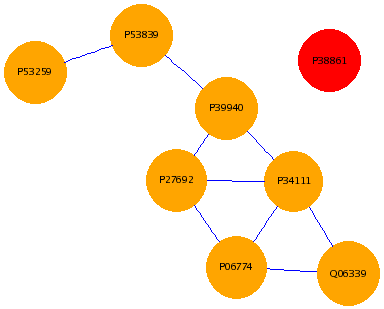
BindingR is a database for Saccharomyces cerevisiae protein-protein interaction binding sites at an aggregate aspect combining domain-domain, domain-motif, and motif-motif levels. It provides users with a web-based interface for fast and convenient access to all potential binding sites of a given interacting protein pair at the three levels.
Proteins usually carry out their functions in a cell by interacting with other molecules such as DNAs, RNAs, and other proteins. Interacting proteins more often contact each other by a domain binding to another domain, by a domain binding to a short protein motif, or by a motif binding to another motif. These predicted binding site pairs at the aggregate level should be helpful for the understanding of the mechanism of protein-protein interactions and be insightful into protein functions, diseases and genomic evolution.
The BindingR database contains the binding sites at the three levels of motif-motif, motif-domain, and domain-domain for Saccharomyces cerevisiae protein-protein interactions. We provide users with four search features for fast and convenient access to binding sites at three levels of protein interactions and potential interacting partner of a domain or motif.

Laboratory of Computational Molecular Biology. Beijing Normal University.