The 6 related genomes
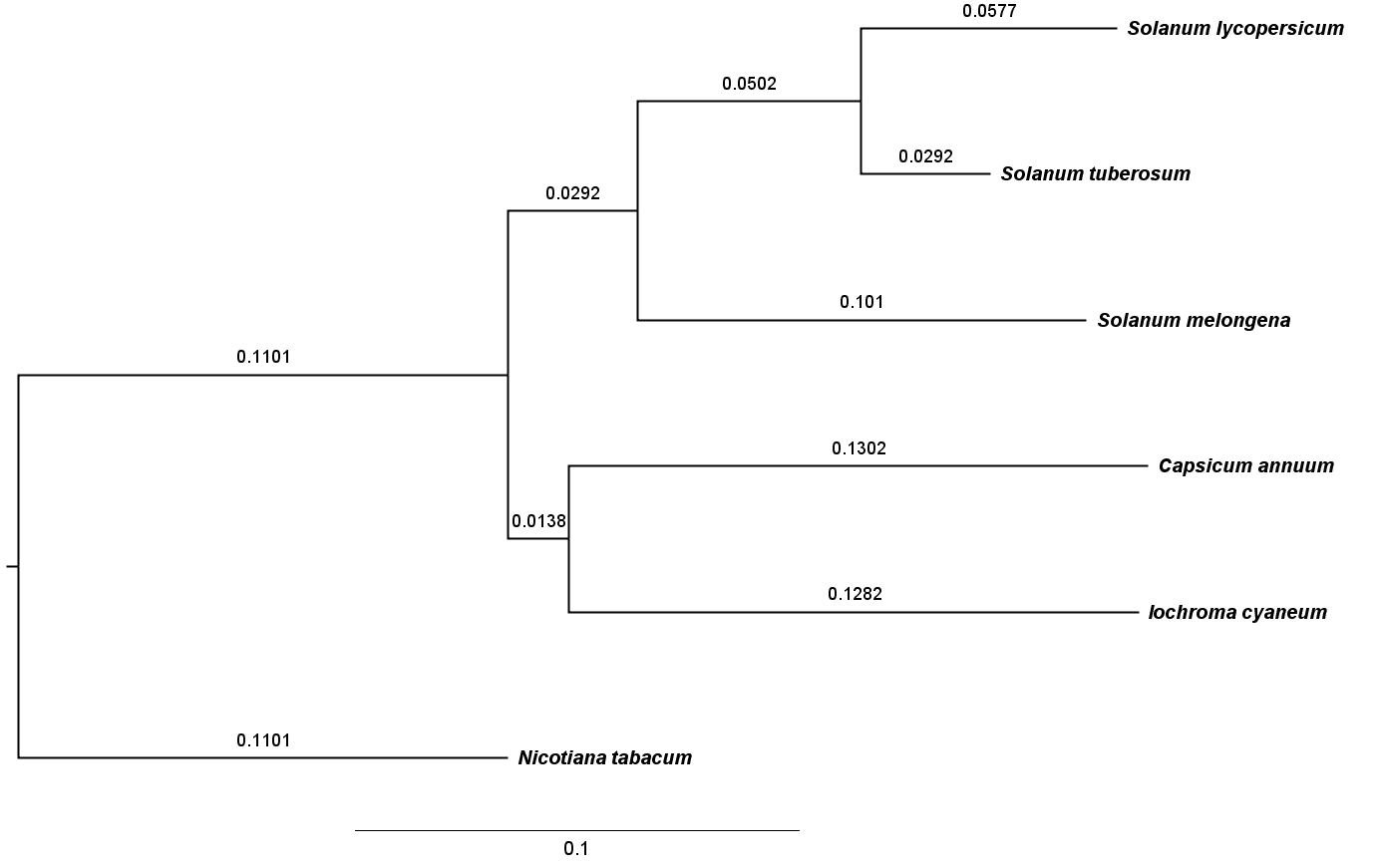
Figure 1. Phylogenetic tree of species included in the 6-way tomato-based alignment,used to guide MULTIZ merging of pairwise alignments
*The neutral tree is based on 4-fold degenerate sites sampled from SL4.0ch01~12 with branch lengths proportional to the indicated scale.
Table 1. Genome data source
| Species | Assembly Name | Annotation Release | Release Date | URL |
|---|---|---|---|---|
| Solanum lycopersicum | SL4.0 | ITAG4.0 | 2019/09/14 | ftp://ftp.solgenomics.net/tomato_genome/assembly/build_4.00/ |
| Solanum tuberosum | V6.1 | V6.1 | 2020/09/23 | http://spuddb.uga.edu/data/ |
| Solanum melongena | V4.1 | V4.1 | 2021/10/18 | https://solgenomics.net/ftp/genomes/Solanum_melongena_V4.1/ |
| Capsicum annuum | V1.0 | V1.0 | 2018/01/25 | https://solgenomics.net/ftp/genomes/Capsicum_annuum/C.annuum_UCD10X/ |
| Iochroma cyaneum | V1.0 | V1.0 | 2021/11/08 | https://solgenomics.net/ftp/genomes/Iochroma_cyaneum/ |
| Nicotiana tabacum | V4.5 | V4.5 | 2017/04/13 | https://solgenomics.net/ftp/genomes/Nicotiana_tabacum/edwards_et_al_2017/assembly/ |