The 7 related genomes used
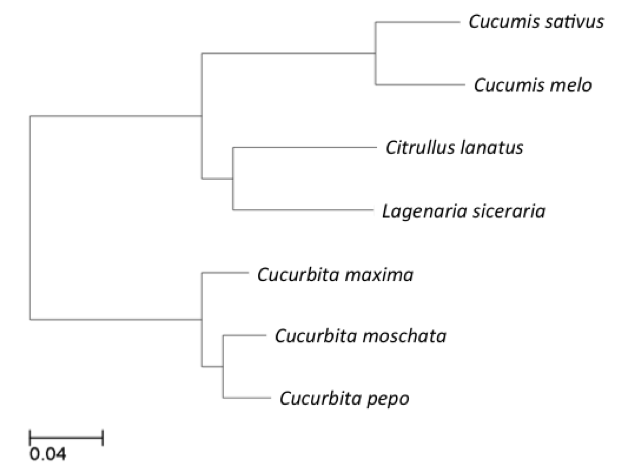
Figure 1. Phylogenetic tree of species included in the 7-way cucumber-based alignment,used to guide MULTIZ merging of pairwise alignments
*The neutral tree is based on 4-fold degenerate sites sampled from Chr1~7 with branch lengths proportional to the indicated scale.The topology of 7 species tree were constructed based 7-way whole genome alignments dataset by UPGMA method
Table 1. Genome data source
| Species | Assembly Name | Annotation Release | Release Date | URL |
|---|---|---|---|---|
| Cucumis sativus | V2.0 | V2 | 2014/08/08 | http://cmb.bnu.edu.cn/Cucumis_sativus_v2.0/index.html |
| Cucumis melo | V3.5.1 | V3.5.1 | 2017/10/16 | ftp://cucurbitgenomics.org/pub/cucurbit/genome/melon/v3.5.1/ |
| Citrullus lanatus | V1 | V1 | 2013/09/21 | ftp://cucurbitgenomics.org/pub/cucurbit/genome/watermelon/97103/ |
| Lagenaria siceraria | V1 | V1 | 2017/10/23 | ftp://cucurbitgenomics.org/pub/cucurbit/genome/Lagenaria_siceraria/ |
| Curcurbita maxima | V1.1 | V1.1 | 2017/09/14 | ftp://ftp.ncbi.nlm.nih.gov/genomes/Malus_domestica |
| Cucurbita moschata | V1 | V1 | 2017/09/14 | ftp://cucurbitgenomics.org/pub/cucurbit/genome/Cucurbita_moschata/ |
| Cucurbita pepo | V4.1 | V4.1 | 2017/11/07 | ftp://cucurbitgenomics.org/pub/cucurbit/genome/Cucurbita_pepo/ |